Detection of binding events from ChIP-exo data¶
Warning
Read the Background Information before proceeding with these steps
Warning
Read the Project Templates Installation notes to have the cookiecutter available in you shell depending on the execution environment you will be using.
Samples description file¶
A TSV file named factors.txt is the main file for the projects and workflow. This file should be created before any project creation. It is the base of the workflow and should be copied to the folder data/{{dataset_name}} just after creating the project structure.
The initial sample names, file name prefixes and metadata are specified on it.
It should have the following columns:
id |
SampleID |
condition |
replicate |
|---|---|---|---|
classical01 |
SRR4053795 |
classical |
1 |
classical01 |
SRR4053796 |
classical |
2 |
nonclassical01 |
SRR4053802 |
nonclassical |
1 |
nonclassical01 |
SRR4053803 |
nonclassical |
2 |
Warning
Columns names are required and are case sensitive.
Columns
id: Sample names. It can be different of sample file name.
SampleID: This is the prefix of the sample file name.
For single-end data the prefix ends in the file extension. In this case, for the first column, a file name named SRR4053795.fastq.gz must exist.
For paired-end data the files SRR4053795_1.fastq.gz and SRR4053795_2.fastq.gz must exist.
The data files should be copied to the folder data/{{dataset_name}}/.
condition: Conditions to analyze or group the samples. Avoid using non alphanumeric characters.
For RNASeq projects the differential gene expression will be generated comparing these conditions. If there are multiple conditions all comparisons will be generated. It must be at least two conditions.
For ChIPSeq projects differential binding events will be detected comparing these conditions. If there are multiple conditions all comparisons will be generated. It must be at least two conditions.
For ChIPexo projects the samples of the same condition will be grouped for the peak calling with MACE.
replicate: Replicate number for samples.
Installation¶
ChIP-exo workflow with Conda/Bioconda¶
The ChIP-exo project structure is created using the conda environment named templates.
First step is to activate the templates environment:
localhost:~> conda activate templates
Then, a YAML file (for this example I will call this file: chipexo-single.yaml) with your project detail should be created.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 | default_context:
author_name: "Roberto Vera Alvarez"
user_email: "veraalva@ncbi.nlm.nih.gov"
project_name: "chipexo-single"
dataset_name: "PRJNA338159"
is_data_in_SRA: "y"
ngs_data_type: "ChIP-exo"
sequencing_technology: "single-end"
create_demo: "n"
number_spots: "1000000"
organism: "human"
genome_dir: "/gfs/data/genomes/NCBI/Escherichia_coli/K-12/MG1655/"
genome_name: "NC_000913.3"
aligner_index_dir: "{{ cookiecutter.genome_dir}}/BWA"
genome_fasta: "{{ cookiecutter.genome_dir}}/NC_000913.3.fa"
genome_gtf: "{{ cookiecutter.genome_dir}}/NC_000913.3.gtf"
genome_gff: "{{ cookiecutter.genome_dir}}/NC_000913.3.gff"
genome_gff3: "{{ cookiecutter.genome_dir}}/NC_000913.3.gff3"
genome_bed: "{{ cookiecutter.genome_dir}}/NC_000913.3.bed"
genome_chromsizes: "{{ cookiecutter.genome_dir}}/NC_000913.3.sizes"
genome_mappable_size: "3714120"
genome_blacklist: "{{ cookiecutter.genome_dir}}/NC_000913.3.bed"
fold_change: "2.0"
fdr: "0.05"
use_docker: "n"
pull_images: "n"
use_conda: "y"
cwl_runner: "cwl-runner"
cwl_workflow_repo: "https://github.com/ncbi/cwl-ngs-workflows-cbb"
create_virtualenv: "n"
use_gnu_parallel: "y"
max_number_threads: "16"
|
A full description of this parameters are here.
After the chipexo-single.yaml is created the project structure should be created using this command obtaining the following output.
localhost:~> cookiecutter --no-input --config-file chipexo-single.yaml https://github.com/ncbi/pm4ngs.git
Checking ChIP-exo workflow dependencies .......... Done
localhost:~>
This process should create a project organizational structure like this:
localhost:~> tree chipexo-single
chipexo-single
├── bin
│ ├── bioconda (This directory include a conda envs for all bioinfo tools)
│ ├── cwl-ngs-workflows-cbb (CWL workflow repo cloned here)
│ └── jupyter (This directory include a conda envs for Jupyter notebooks)
├── config
│ └── init.py
├── data
│ └── PRJNA338159
├── index.html
├── LICENSE
├── notebooks
│ ├── 00 - Project Report.ipynb
│ ├── 01 - Pre-processing QC.ipynb
│ ├── 02 - Samples trimming.ipynb
│ ├── 03 - Alignments.ipynb
│ ├── 04 - Peak Calling.ipynb
│ └── 05 - MEME Motif.ipynb
├── README.md
├── requirements
│ └── python.txt
├── results
│ └── PRJNA338159
├── src
└── tmp
10 directories, 11 files
Now you should copied the factors.txt file to the folder: data/PRJNA338159.
After this process, cookiecutter should have created create two virtual environment for this workflow.
The first one is for running the Jupyter notebooks which require Python 3.6+ and it is named: jupyter. It can be manually installed as described in here.
The second environment is be used to install all Bioinformatics tools required by the workflow and it will be named: bioconda.
You can verify the environments running this command:
localhost:~> conda env list
# conda environments:
#
base * /gfs/conda
tempates /gfs/conda/envs/templates
/home/veraalva/chipexo-single/bin/bioconda
/home/veraalva/chipexo-single/bin/jupyter
localhost:~>
Please, note that the Conda prefix /gfs/conda will be different in you host. Also, note that the bioconda and jupyter envs are inside the bin directory of your project keeping them static inside the project organizational structure.
ChIP-exo workflow usage with Conda/Bioconda¶
For start using the workflow you need to activate the conda environments bioconda and jupyter.
localhost:~> conda activate /home/veraalva/chipexo-single/bin/bioconda
localhost:~> conda activate --stack /home/veraalva/chipexo-single/bin/jupyter
Note the --stack option to have both environment working at the same time. Also, the order is important, bioconda should be activated before jupyter.
Test the conda envs:
localhost:~> which fastqc
/home/veraalva/chipexo-single/bin/bioconda/bin/fastqc
localhost:~> which jupyter
/home/veraalva/chipexo-single/bin/jupyter/bin/jupyter
Note that the fastqc tools is installed in the bioconda env and the jupyter command is installed in the jupyter env.
Then, you can start the jupyter notebooks.
localhost:~> jupyter notebook
If the workflow is deployed in a remote machine using SSH access the correct way to start the notebooks is:
localhost:~> jupyter notebook --no-browser --ip='0.0.0.0'
In this case the option --ip='0.0.0.0' will server the Jupyter notebook on all network interfaces and you can access them from your desktop browser using the port returned by the Jupyter server.
Finally, you should navegate in your browser to the notebooks folder and start executing all notebooks by their order leaving the 00 - Project Report.ipynb to the end.
ChIP-exo workflow with Docker¶
In this case, the ChIP-exo project structure is created using the Python virtual environment as described here
First step is to activate the Python virtual environment.
localhost:~> source venv-templates/bin/activate
Then, a YAML file (for this example I will call this file: chipexo-single.yaml) with your project detail should be created.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 | default_context:
author_name: "Roberto Vera Alvarez"
user_email: "veraalva@ncbi.nlm.nih.gov"
project_name: "chipexo-single"
dataset_name: "PRJNA338159"
is_data_in_SRA: "y"
ngs_data_type: "ChIP-exo"
sequencing_technology: "single-end"
create_demo: "n"
number_spots: "1000000"
organism: "human"
genome_dir: "/gfs/data/genomes/NCBI/Escherichia_coli/K-12/MG1655/"
genome_name: "NC_000913.3"
aligner_index_dir: "{{ cookiecutter.genome_dir}}/BWA"
genome_fasta: "{{ cookiecutter.genome_dir}}/NC_000913.3.fa"
genome_gtf: "{{ cookiecutter.genome_dir}}/NC_000913.3.gtf"
genome_gff: "{{ cookiecutter.genome_dir}}/NC_000913.3.gff"
genome_gff3: "{{ cookiecutter.genome_dir}}/NC_000913.3.gff3"
genome_bed: "{{ cookiecutter.genome_dir}}/NC_000913.3.bed"
genome_chromsizes: "{{ cookiecutter.genome_dir}}/NC_000913.3.sizes"
genome_mappable_size: "3714120"
genome_blacklist: "{{ cookiecutter.genome_dir}}/NC_000913.3.bed"
fold_change: "2.0"
fdr: "0.05"
use_docker: "y"
pull_images: "y"
use_conda: "n"
cwl_runner: "cwl-runner"
cwl_workflow_repo: "https://github.com/ncbi/cwl-ngs-workflows-cbb"
create_virtualenv: "y"
use_gnu_parallel: "y"
max_number_threads: "16"
|
A full description of this parameters are here.
After the chipexo-single.yaml is created the project structure should be created using this command obtaining the following output.
localhost:~> cookiecutter --no-input --config-file chipexo-single.yaml https://github.com/ncbi/pm4ngs.git
Checking ChIP-exo workflow dependencies .......... Done
localhost:~>
This process should create a project organizational structure like this:
localhost:~> tree chipexo-single
chipexo-single
├── bin
├── config
│ └── init.py
├── data
│ └── PRJNA338159
├── index.html
├── LICENSE
├── notebooks
│ ├── 00 - Project Report.ipynb
│ ├── 01 - Pre-processing QC.ipynb
│ ├── 02 - Samples trimming.ipynb
│ ├── 03 - Alignments.ipynb
│ ├── 04 - Peak Calling.ipynb
│ └── 05 - MEME Motif.ipynb
├── README.md
├── requirements
│ └── python.txt
├── results
│ └── PRJNA338159
├── src
├── tmp
└── venv
11 directories, 11 files
Now you should copied the factors.txt file to the directory: data/PRJNA338159.
After this process, cookiecutter should have pulled all docker images require by the project.
ChIP-exo workflow usage with Docker¶
For start using the workflow you need to activate the Python environment inside the project.
localhost:~> source venv/bin/activate
Then, you can start the jupyter notebooks now.
localhost:~> jupyter notebook
If the workflow is deployed in a remote machine using SSH access the correct way to start the notebooks is:
localhost:~> jupyter notebook --no-browser --ip='0.0.0.0'
In this case the option --ip='0.0.0.0' will server the Jupyter notebook on all network interfaces and you can access them from your desktop browser using the port returned by the Jupyter server.
Finally, you should navigate in your browser to the notebooks directory and start executing all notebooks by their order leaving the 00 - Project Report.ipynb to the end.
Jupyter Notebook Server¶
Top-level directories from the Jupyter server viewed in a web browser¶
Notebook generated fro the Chip-exo data analysis¶
CWL workflows¶
SRA download and QC workflow¶
This CWL workflow is designed to download FASTQ files from the NCBI SRA database using fastq-dump and then, execute fastqc generating a quality control report of the sample.
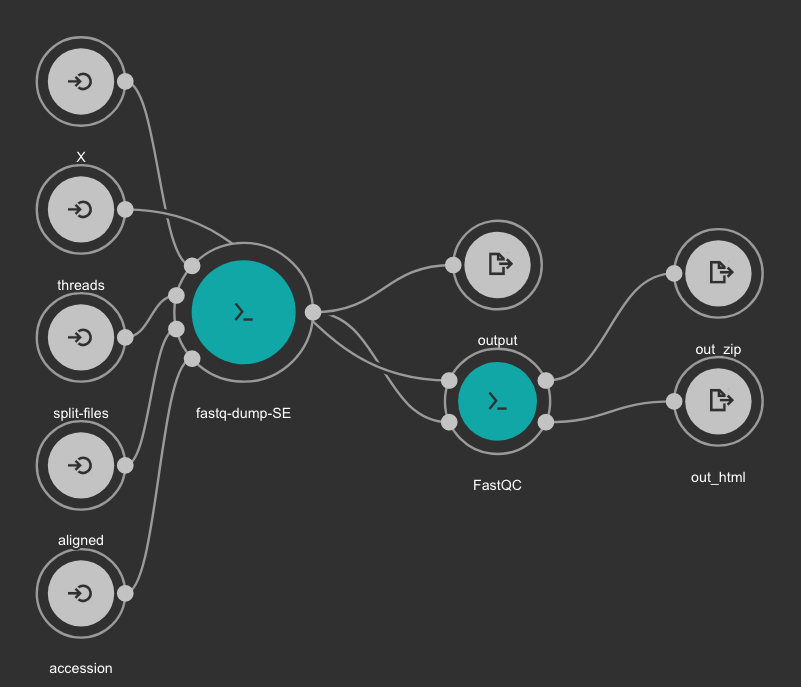
Inputs
accession: SRA accession ID. Type: string. Required.
aligned: Used it to download only aligned reads. Type: boolean. Optional.
split-files: Dump each read into separate file. Files will receive suffix corresponding to read number. Type: boolean. Optional.
threads: Number of threads. Type: int. Default: 1. Optional.
X: Maximum spot id. Optional.
Outputs
output: Fastq files downloaded. Type: File[]
out_zip: FastQC report ZIP file. Type: File[]
out_html: FastQC report HTML. Type: File[]
Samples Trimming¶
Our workflows uses Trimmomatic for read trimming. The Jupyter notebooks uses some basic Trimmomatic options that need to be modified depending on the FastQC quality control report generated for the sample.
BWA based alignment and quality control workflow¶
This workflow use BWA as base aligner. It also use SAMtools and bedtools for file conversion and statistics report. Finally, Phantompeakqualtools is used to generate quality control report for the processed samples.
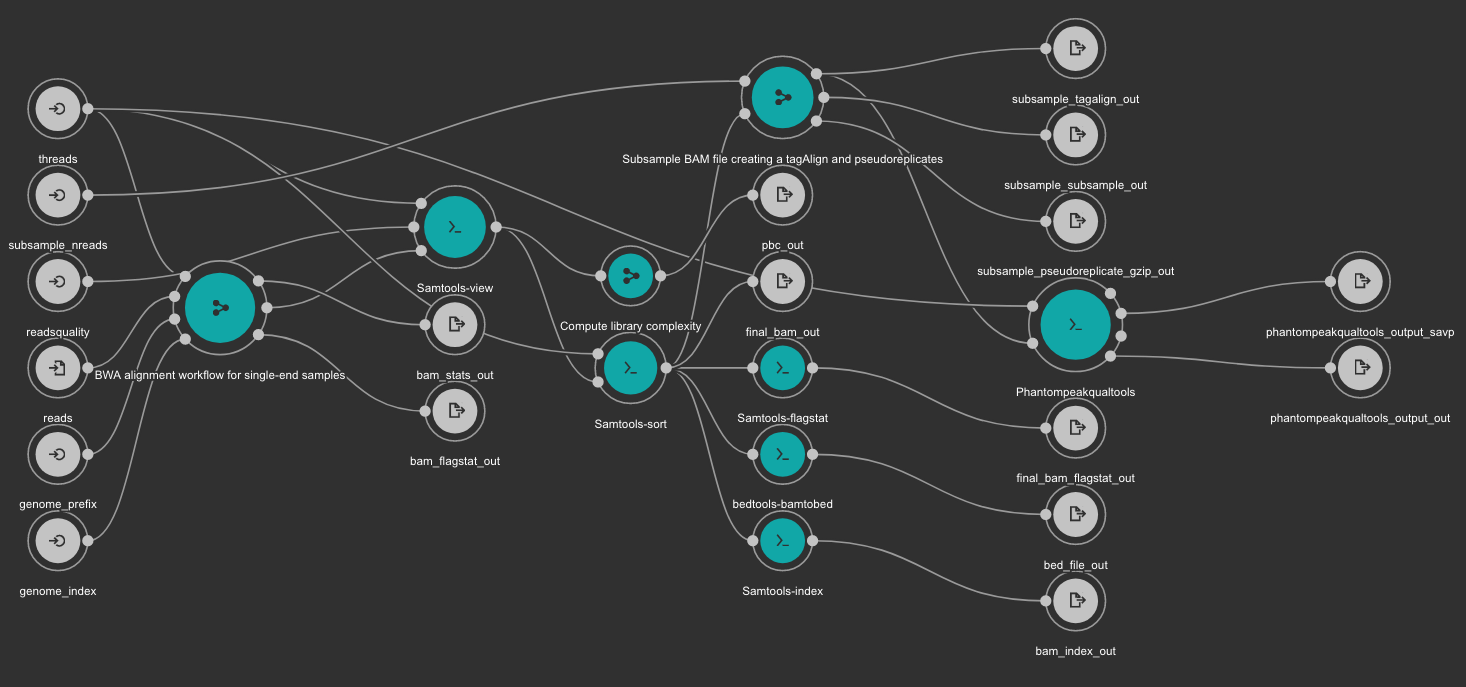
Inputs
genome_index: Aligner indexes directory. Type: Directory. Required. Variable ALIGNER_INDEX in the Jupyter Notebooks.
genome_prefix: Prefix of the aligner indexes. Generally, it is the name of the genome FASTA file. It can be used as os.path.basename(GENOME_FASTA) in the Jupyter Notebooks. Type: string. Required.
readsquality: Minimum MAPQ value to use reads. We recommend for ChIP_exo dataa value of: 30. Type: int. Required.
threads: Number of threads. Type: int. Default: 1. Optional.
subsample_nreads: Number of reads to be used for the subsample. We recomend for ChIP_exo dataa value of: 500000. Type: int. Required.
reads: FastQ file to be processed. Type: File. Required.
Outputs
bam_flagstat_out: SAMtools flagstats for unfiltered BAM file. Type: File.
bam_stats_out: SAMtools stats for unfiltered BAM file. Type: File.
final_bam_flagstat_out: SAMtools flagstats for filtered BAM file. Type: File.
bed_file_out:: Aligned reads in BED format. Type: File.
final_bam_out: Final BAM file filtered and sorted. Type: File.
bam_index_out: BAM index file. Type: File.
pbc_out: Library complexity report. Type: File.
phantompeakqualtools_output_out: Phantompeakqualtools main output. Type: File.
phantompeakqualtools_output_savp: Phantompeakqualtools SAVP output. Type: File.
subsample_pseudoreplicate_gzip_out: Subsample pseudoreplicates tagAlign gzipped. Type: File[].
subsample_tagalign_out: Subsample tagAlign gzipped. Type: File[].
subsample_subsample_out: Subsample shuffled tagAlign gzipped. Type: File[].
Peak caller workflow using MACE¶
This workflow uses MACE as peak caller tool. The annotation is created from the GTF file using a in-house python script available here. BAMscale is used for the quantification of the resulting peaks.
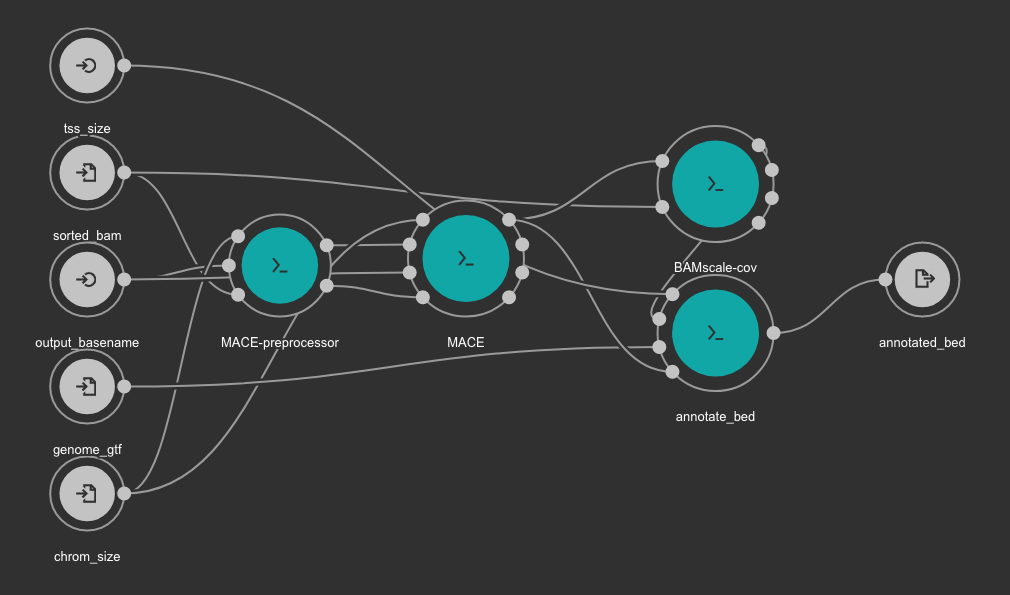
Inputs
chrom_size: Chromosome size file. Tab or space separated text file with 2 columns: first column is chromosome name, second column is size of the chromosome. Type: File. Required. Variable GENOME_CHROMSIZES in the Jupyter Notebooks.
output_basename: Prefix for the output file. Type: string. Required.
genome_gtf: Genome GTF file. Variable GENOME_GTF in the Jupyter Notebooks. Type: File. Required.
tss_size: Number of bp to use for TSS definition. We recommend use 1000. Type: int. Required.
Outputs
annotated_bed: Annotated detected peaks in BED format. Type: File
MEME Motif detection workflow¶
Motif detection is executed using the MEME suite.
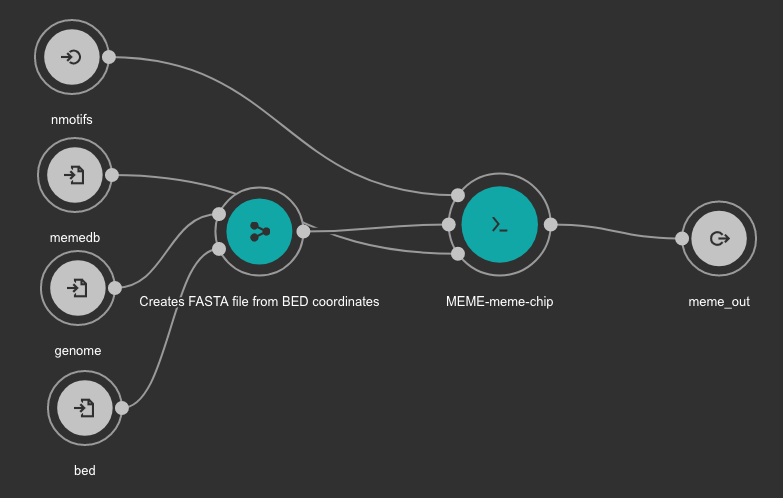
Inputs
bed: BED file with detected peaks. Type: File. Required.
memedb: MEME database for use by Tomtom and CentriMo. Type: File. Required.
genome: Genome FASTA file. Variable GENOME_FASTA in the Jupyter Notebooks. Type: File. Required.
nmotifs: Maximum number of motifs to find. We recommend use 10. Type: int. Required.
Outputs
meme_out: MEME output directory. Type: Directory
MEME databases¶
MEME workflow depends on the MEME databases. Go to the MEME Suite Download page: http://meme-suite.org/doc/download.html
Download the latest version for the Motif Databases and GOMo Databases.
The downloaded files should be uncompressed in a directory data/meme. The final directory should be:
localhost:~> cd data
localhost:~> mkdir meme
localhost:~> cd meme
localhost:~> wget http://meme-suite.org/meme-software/Databases/motifs/motif_databases.12.18.tgz
localhost:~> wget http://meme-suite.org/meme-software/Databases/gomo/gomo_databases.3.2.tgz
localhost:~> tar xzf motif_databases.12.18.tgz
localhost:~> tar xzf gomo_databases.3.2.tgz
localhost:~> rm gomo_databases.3.2.tgz motif_databases.12.18.tgz
localhost:~> tree -d
.
├── gomo_databases
└── motif_databases
├── ARABD
├── CIS-BP
├── CISBP-RNA
├── ECOLI
├── EUKARYOTE
├── FLY
├── HUMAN
├── JASPAR
├── MALARIA
├── MIRBASE
├── MOUSE
├── PROKARYOTE
├── PROTEIN
├── RNA
├── TFBSshape
├── WORM
└── YEAST
19 directories
See also an example in our test project: https://ftp.ncbi.nlm.nih.gov/pub/pm4ngs/examples/chipexo-single/data/
Test Project¶
A test project is available (read-only) at https://ftp.ncbi.nlm.nih.gov/pub/pm4ngs/examples/chipexo-single/
Extra requirements¶
Creating BWA indexes¶
This workflow uses BWA for sequence alignment. The BWA index creation is not included in the workflow, that's why we are including an small section here to describe how the BWA indexes can be created.
The genome.fa file should be copied to the genome directory.
localhost:~> conda activate /home/veraalva/chipexo-single/bin/bioconda
localhost:~> conda activate --stack /home/veraalva/chipexo-single/bin/jupyter
localhost:~> cd chipexo-single/data
localhost:~> mkdir genome
localhost:~> cd genome
localhost:~> mkdir BWA
localhost:~> cd BWA
localhost:~> cwl-runner --no-container ../../../bin/cwl-ngs-workflows-cbb/tools/BWA/bwa-index.cwl --sequences genome.fa
localhost:~> cd ..
localhost:~> tree
.
├── BWA
│ ├── genome.fa
│ ├── genome.fa.amb
│ ├── genome.fa.ann
│ ├── genome.fa.bwt
│ ├── genome.fa.pac
│ └── genome.fa.sa
└── genome.fa
1 directory, 7 files
Here all files inside the directory BWA are created by the workflow.
Creating BED files from GTF¶
For generating a BED file from a GTF.
The genes.gtf file should be copied to the genome directory.
localhost:~> conda activate /home/veraalva/chipexo-single/bin/bioconda
localhost:~> conda activate --stack /home/veraalva/chipexo-single/bin/jupyter
localhost:~> cd chipexo-single/data
localhost:~> mkdir genome
localhost:~> cd genome
localhost:~> cwl-runner --no-container ../../bin/cwl-ngs-workflows-cbb/workflows/UCSC/gtftobed.cwl --gtf genes.gtf
localhost:~> tree
.
├── genes.bed
├── genes.genePred
├── genes.gtf
└── genome.fa
0 directory, 4 files
Here the files genes.bed and genes.genePred are created from the workflow.